Development of Retrotransposons Into Practical Molecular Markers
Molecular, or DNA-based, markers have been increasingly important in plant breeding because of their phenotypic stability, useful polymorphism, and ease of development which permits production of high-density maps for use in marker-assisted breeding. These have proven superior to both biochemical (protein) and phenotypic markers, which suffer from a fairly low degree of polymorphism, lack of sufficient loci to generate dense maps, and environmentally variable expression. To generate a multiplicity of bands and maximize the potential to detect genetic polymorphisms, all molecular marker systems except RFLP (restriction fragment length polymorphism) are based on prevalent, widespread elements in the genome. Most of these systems rely of PCR amplification, either from random primers (
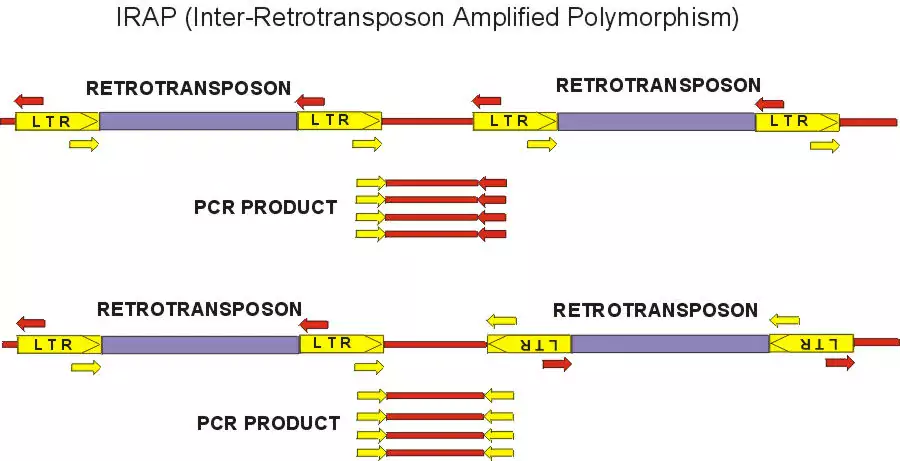
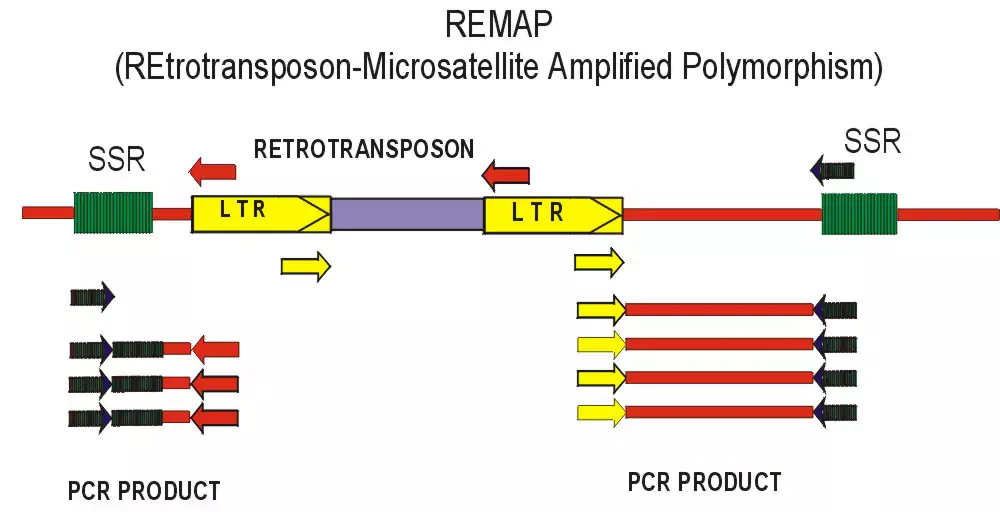
Retrotransposons can be used for markers because integration of a daughter copy creates new joints between genomic DNA and the conserved LTRs. To detect polymorphisms for retrotransposon insertion, the marker systems that have been recently developed generally rely on PCR amplification between an LTR and some component of the flanking genomic DNA. We have developed two methods, REMAP (REtotransposon-Microsatellite Amplified Polymorphism) and IRAP (Inter-Retrotransposon Amplified Polymorphism) which require neither restriction enzyme digestion nor ligation to generate the marker bands. The IRAP bands are generated from two nearby LTRs using outward-facing primers annealing to LTR target sequences. In REMAP, amplification between LTRs proximal to simple sequence repeats such microsatellites produces the marker bands.
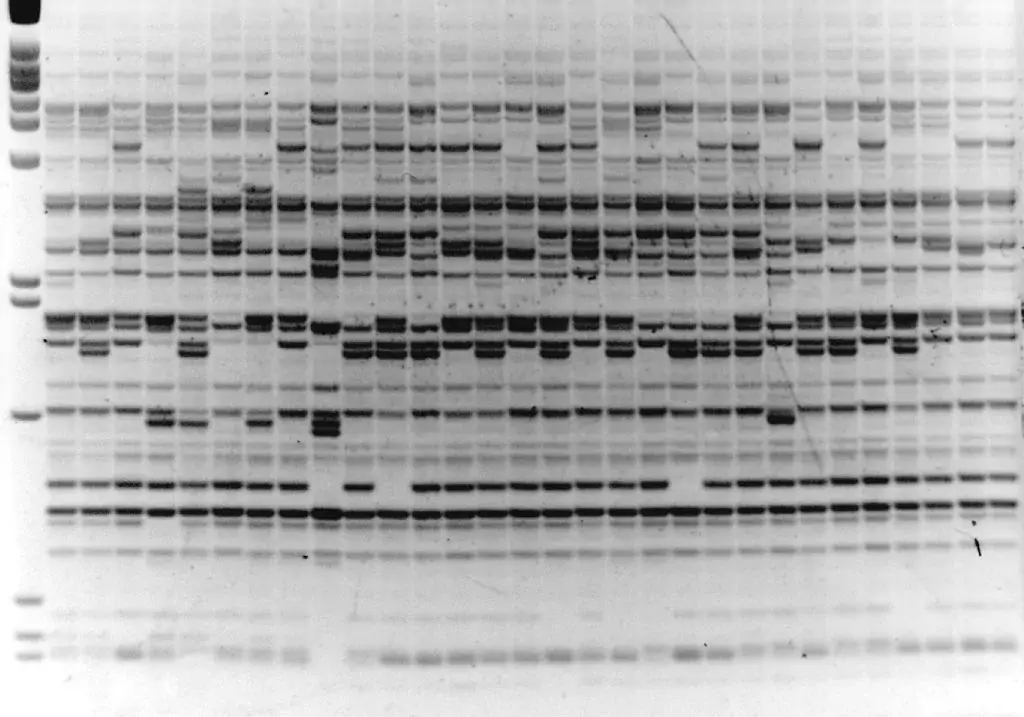
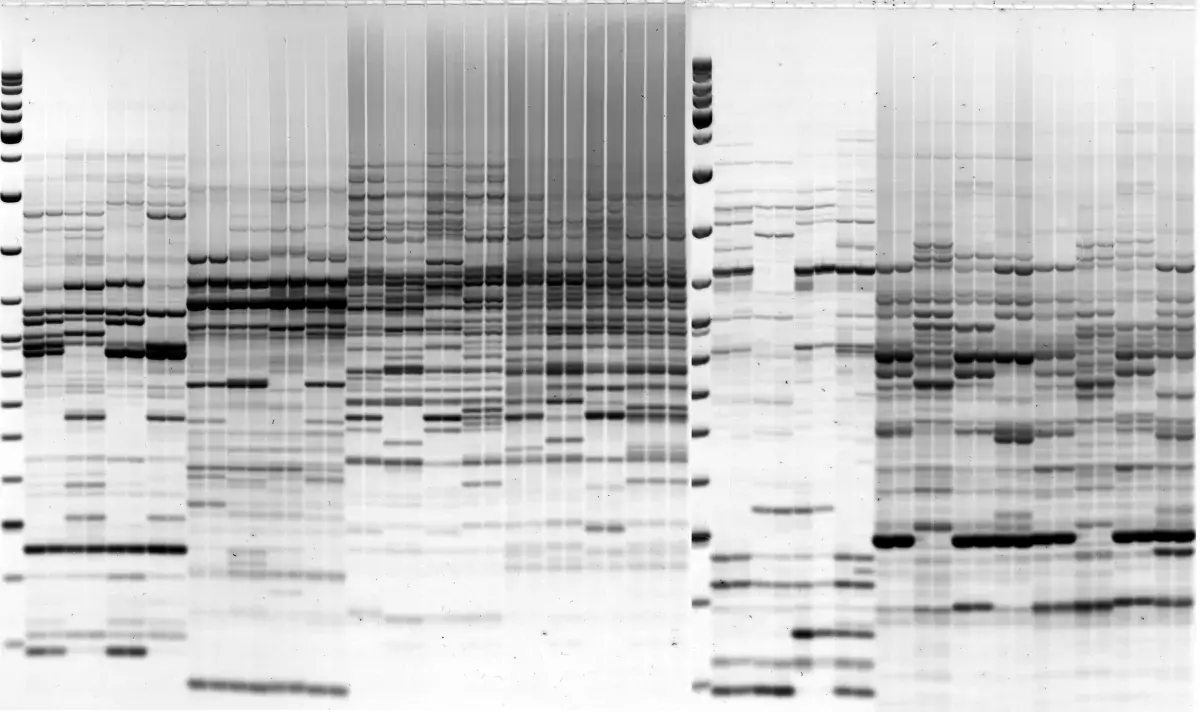
Image 1: IRAP with Sukkula LTR primer, barley varieties
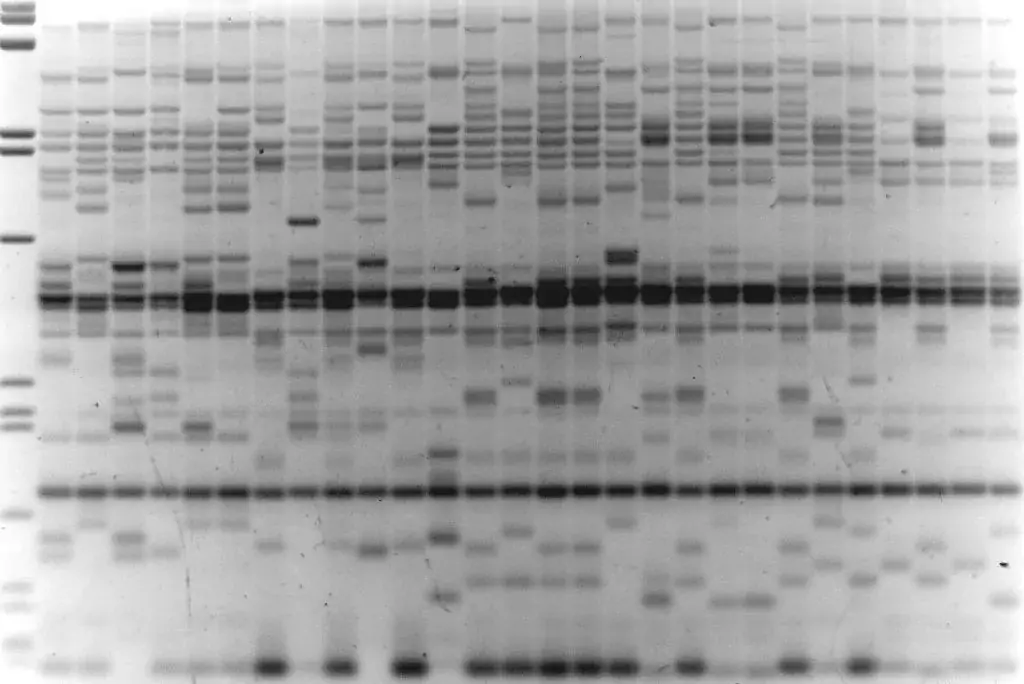
Image 2: REMAP with BARE1 LTR primer, H.spontaneum individuals
Retrotransposons have several advantages as molecular markers over the other systems described above. Their abundance and dispersion can yield many marker bands, the pattern possessing a high degree of polymorphism due to transpositional activity. The LTR termini are highly conserved even between families, yet longer primers can be tailored to specific families. Unlike DNA transposons, the new copies are inserted but not removed. Even intra-element recombination resulting in the conversion of a full-length element to a solo LTR either does not affect the marker band and can furthermore be exploited to produce a new marker band in RBIP. Retrotransposon families may vary in their insertional activity, as is seen clearly by the pattern of nested insertions in maize, allowing the matching of the family used for marker generation to the phylogenetic depth required. Furthermore, the length and conservation of primers to the LTRs facilitate cloning of interesting marker bands and the development of new retrotransposons for markers.
IRAP, REMAP and iPBS for retrotransposon-based genotyping and fingerprinting protocols
The long terminal repeat (LTR) retrotransposons are well suited as molecular markers. Both the overall structure of retrotransposons and the domains responsible for the various phases of their replication are highly conserved in all eukaryotes. Nevertheless, up to a year has been required to develop a retrotransposon marker system in a new species, involving cloning and sequencing steps as well as the development of custom primers.
We describe a novel PCR-based method useful both as a marker system in its own right and for the rapid isolation of retrotransposon termini and full-length elements, making it ideal for "orphan crops" and other species with underdeveloped marker systems.
The method, iPBS amplification, is based on the virtually universal presence of a tRNA complement as a reverse transcriptase primer binding site (PBS) in LTR retrotransposons. It is based on the nearly universal use by both retroviruses and LTR retrotransposons of cellular tRNAs as primers for reverse transcription during their replication cycles. The tRNA binds to the primer binding site (PBS) adjacent to the 5' LTR and primes synthesis of minus-strand cDNA by reverse transcriptase. A handful of exceptional LTR retrotransposons, including the Tf1/sushi group of fungi and vertebrates and Fourf in maize, is able to self-prime cDNA synthesis rather than use a tRNA primer.
The method differs from earlier retrotransposon isolation methods because it is applicable not only to endogenous retroviruses and retroviruses, but also to both Gypsy and Copia LTR retrotransposons, as well as to non-autonomous LARD and TRIM elements, throughout the plant kingdom and to animals. Furthermore, the inter-PBS amplification technique as such has proved to be a powerful DNA fingerprinting technology without the need for prior sequence knowledge.
The use of the PBS offers several advantages
First, it is universal, in that virtually all retrotransposons, including TRIMs and LARDs, which do not have internal coding domains, serve as targets.
Second, it can be used both for cloning retrotransposons and ERVs as well as for displaying their insertion sites. Other methods, relying on conserved protein coding domains of retrotransposons, require walking to the LTRs in order to produce a marker system. Third, the PBS is adjacent to the LTR, facilitating its isolation.
Last, a fairly small set of primers, once produced, can be used in any organism with a complement of matching elements. These advantages offer savings of time and money in the development of new marker systems for "orphan crops," wild species, and other species for which resources are limited or for which the development of SNP-based markers is impractical. The simple analytical systems for retrotransposon markers only strengthen this benefit.
We show that using a full set of the conserved parts of PBS sequences, we are able both to directly visualize polymorphism between individuals, rapidly clone LTR segments from genomic DNA, and carry out in silico database searches. The method is applicable to any organism with retrotransposons containing PBS sites complimentary to tRNA.
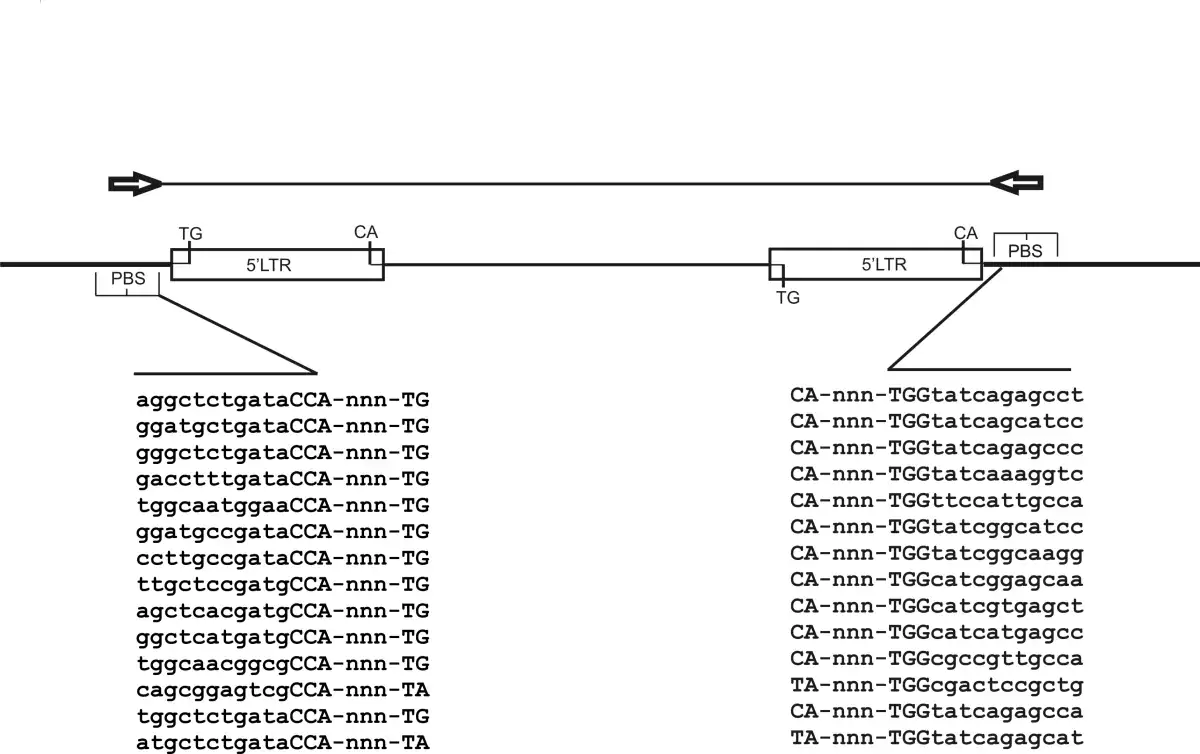
The iPBS scheme. For iPBS, two retrotransposons must be in opposite orientation and either near enough for efficient amplification, as shown, or nested. The diagram depicts two key structural features of retrotransposons, the LTR (long terminal repeat) and PBS (primer binding site). The internal domain is shown as a thick bar, the intervening genomic DNA as thick line. The predicted product is show above, together with the orientation of the PBS amplification primers. The PCR product contains both LTRs and PBS sequences together with the genomic sequence between the LTRs. Below the diagram The sequence of a set of PBS domains, the 0-5 base spacer and the universal 5' TG of LTRs is shown.
Image 1: The iPBS scheme
Image 2: iPBS retrotransposon-based genotyping and fingerprinting protocols
Publications:
Kalendar R, Schulman AH 2006.
Kalendar R, Antonius K, Smykal P, Schulman AH 2010.
Kalendar R 2011. The use of retrotransposon-based molecular markers to analyze genetic diversity. Field and Vegetable Crops Research, 48(2): 261-274. DOI:10.5937/ratpov1102261K
Schulman A, Flavell A, Paux E, Noel Ellis TH 2012.
Kalendar R, Schulman AH 2014.
Kalendar R, Aizharkyn KS, Khapilina ON, Amenov AA, Tagimanova DS 2017.
Kalendar R, Amenov A, Daniyarov A 2019.